Practical exercises – Bioinformatics teachings (Olivier Croce)
First steps for assembling reads : use of CAP3 software
There are a huge range of assemblers program, with various specificity. CAP3 is one of the older and is only few in use today ,because of its speed and efficiency. However, it is roughly simple to use.
Suppose that you are already connect to the bioserver in ssh mode as described in CLC tutorial. Go into the directory « formation_bioinfo » using command « cd » :
![]()
You can display the content of this folder using « ls -l »
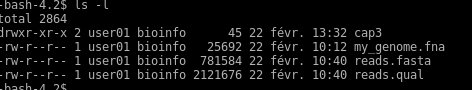
The first step should be to create your own unique folder as you share the same ssh account with other students. Use « mkdir » command with you name for example :
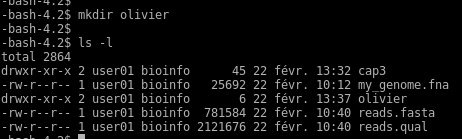
If you type « ls -l » the new directory you created will appear in list
Ok, now go inside your directory :
![]()
And type the following command :
![]()
The assembly process could take around 1 minute for such a set of reads (the same as we used in CLC).
When is done, some files were generated, which you can see using « ls -l » command :

You can have a look of the content of each file using linux commands such a « less » or « more » (i.e. « less reads.fasta.cap.contigs »)
If you wish to play with CAP3 by changing options, here the list of options :
Options (default values):
-a N specify band expansion size N > 10 (20)
-b N specify base quality cutoff for differences N > 15 (20)
-c N specify base quality cutoff for clipping N > 5 (10)
-d N specify max qscore sum at differences N > 100 (250)
-e N specify extra number of differences N > 10 (20)
-g N specify gap penalty factor N > 0 (6)
-m N specify match score factor N > 0 (2)
-n N specify mismatch score factor N < 0 (-5)
-o N specify overlap length cutoff > 20 (30)
-p N specify overlap percent identity cutoff N > 65 (75)
-s N specify overlap similarity score cutoff N > 100 (500)
-u N specify min number of constraints for correction N > 0 (4)
-v N specify min number of constraints for linking N > 0 (2)
-x N specify prefix string for output file names (cap)
If no quality file is given, then a default quality value of 10 is used for each base.