User Tools
Sidebar
This is an old revision of the document!
16S Microbial Analysis (with Galaxy)
This tutorial required:
- a computer (under Linux, Mac, Windows) with an internet connection
- To know what a fasta or fastq (fastaq = fq) file is
Click here for the fasta format description
Click here for the fasta format description
Data in text format used to store nucleic acid sequences (such as DNA sequences) or protein sequences; may contain multiple sequences. FASTA files often start with a header line that may contain comments or other information. The rest of the file contains sequence data. Each sequence starts with a “>” symbol followed by the name of the sequence. The rest of the line describes the sequence and the remaining lines contain the sequence itself.
Example of a fasta file including 2 sequences:
>human_T1 (UCSC April 2002 chr7:115977709:)
TTGTCAGATTCACCAAAGTTGAAATGAAGGAAAAAATGCTAAGGGCAGCC
AGAGAGAGGTCAGGTTACCCACAAAGGGAAGCCCATCAGACTAACAGCG
ATCTCTCGGCAGAAACCCTACAGGCCAGAAGAGAGTGGGGGCCAATATT
CATATTCTTAAAGAAAAGAATTTTCAACCCAGAATTTCATATCCAGCCAA
>human_T2 (UCSC April 2002 chr5:11977710:)
ATACGACYCTTATTGTTAGTATATAATTTATATGAAAACMAAAAATTATG
GCGGTATTTTAAGCTTTTCAGAGGAATTTGCTCTTTAATGGATAAAAC
CTAAATCTTACTAGAATTAGTAAAGCAGTTTGTATACCACT
- an access to a Galaxy website server, including usual tools for metagenomic analysis
See what is the Galaxy project: Wikipedia or Official website
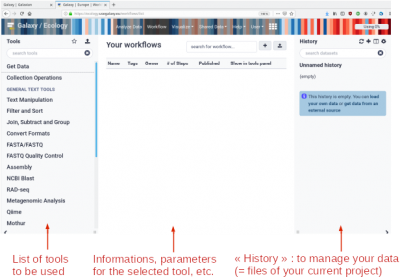 Example of the main display page of Galaxy (using a browser)
Example of the main display page of Galaxy (using a browser)
⇒ 3 possibilities to get access to a Galaxy server:
1) Use a free distant server
Many public Galaxy servers are available. Here a list of servers including the required metaG tools (list updated November 2022):
https://africa.usegalaxy.eu/
https://india.usegalaxy.eu/
https://ecology.usegalaxy.eu/
https://galaxy.genouest.org/
https://metagenomics.usegalaxy.eu/
https://streetscience.usegalaxy.eu/
https://usegalaxy.be/
https://usegalaxy.eu/
https://usegalaxy.fr/
https://usegalaxy.no/
https://usegalaxy.org/
https://usegalaxy.org.au/
pro: no installation required, usable directly
cons: some limitations and slow (or super slow) when running big computations
2) Use our local Galaxy server (IRCAN institute, in Pasteur tower)
http://galaxy-metag.ircan.org/
pro: no installation required, usable directly
cons: should to be faster than previous servers, but depending the number of students connected at the same time !
3) Install Galaxy on your own computer
- install “docker” tool here
- install a Galaxy server into Docker and run docker, by typing this single command:
docker run -p 8080:80 quay.io/galaxy/metagenomics-training
Wait a loooong time before it starts : 5-10 Go to download the first time! ⇒ do it at home using a fast internet connection. Further starts are faster (re type the same command to enable the server).
- Go here in your browser : http://localhost:8080
pro: fast server, depending of you computer power. Reusable.
cons: harder (and long) to install. Take some disk space onto your hard drive.
then,
- Create a personal account into Galaxy (top-right menu)
- Start this tutorial : Here (alternative link)
